Abstract
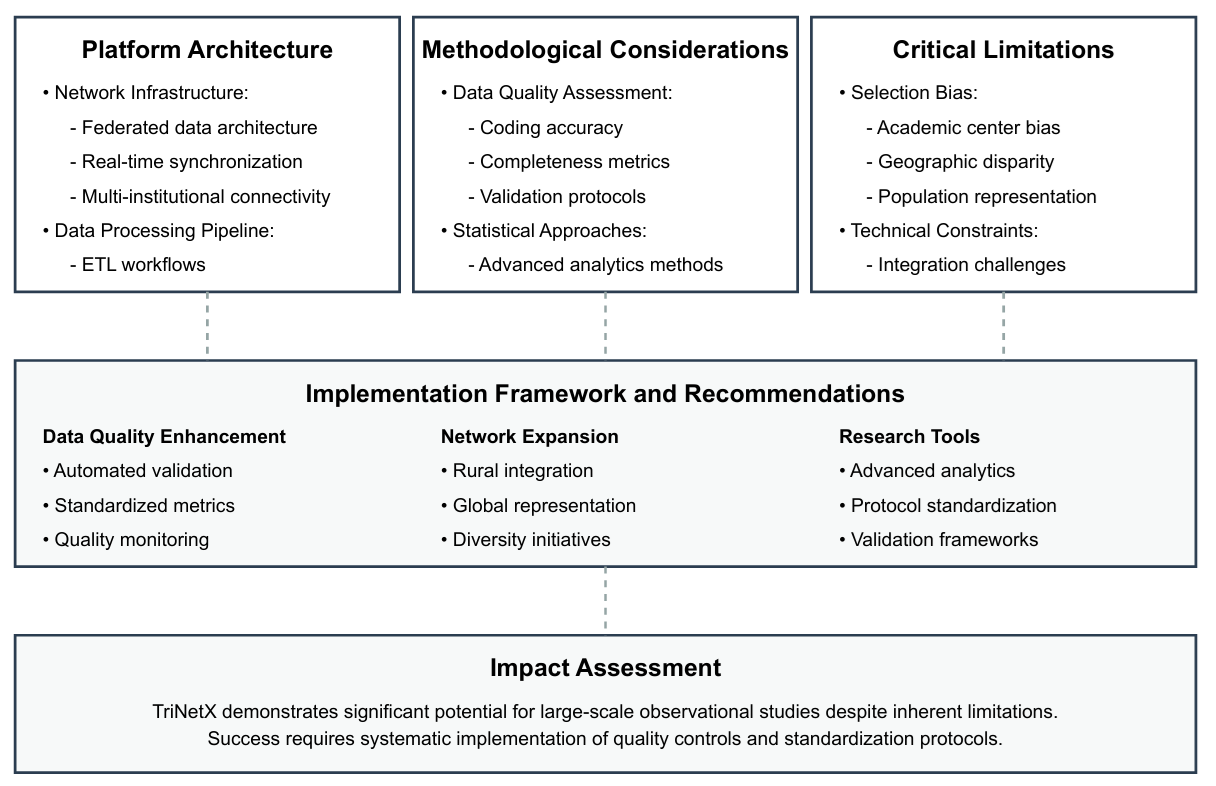
Introduction: The increasing utilization of real-world data platforms in medical research necessitates a comprehensive understanding of their methodological strengths and limitations. TriNetX has emerged as a significant platform for exploring large healthcare datasets. This review aims to critically evaluate the methodological framework and limitations of TriNetX, assess the impact of electronic health record coding accuracy on data reliability, and analyze the platform's capacity for generating generalizable real-world evidence in clinical research.
Methods: We conducted a comprehensive review examining TriNetX's data architecture, quality metrics, and research applications, focusing on data integrity, platform architecture, and the external validity of research findings.
Results: The analysis reveals significant methodological considerations. TriNetX's reliance on retrospective data introduces biases such as selection bias and confounding variables. The coding accuracy of electronic health records, which have not been independently validated, is a critical determinant of data reliability. The demographic representation is limited, affecting the generalizability of results.
Discussion: Despite its extensive use, TriNetX's effective utilization requires careful consideration of its inherent limitations. The platform's data, predominantly from insured populations in academic and acute care settings, may not fully represent broader demographic groups. Addressing these methodological constraints is crucial for enhancing the reliability and applicability of research findings derived from TriNetX.
Conclusions: TriNetX is a valuable resource for healthcare research. However, its limitations must be acknowledged, and future research should focus on standardizing data collection and enhancing data validation processes to mitigate platform-specific biases and improve the quality and applicability of the findings.
References
1. Palchuk MB, London JW, Perez-Rey D, Drebert ZJ, Winer-Jones JP, Thompson CN, Esposito J, Claerhout B. A global federated real-world data and analytics platform for research. JAMIA Open. 2023: ooad035 [PMID: 37193038 https://doi.org/10.1093/jamiaopen/ooad035]
2. Sáez Marín AJ, Hernández-Ibarburu G, Alonso Fernández R, Sanchez Pina JM, Lázaro Del Campo P, Jimenez Ubieto A, Cuellar C, Tamayo Soto A, Mas Babio R, Medina L, Calbacho M, Ayala R, Martinez Lopez J. Large Scale Analysis of Autologous Stem Cell Transplantation for Multiple Myeloma Patients Older Than 65 Years. Blood. 2023: 7064 https://doi.org/10.1182/blood-2023-180923]
3. Singh D, Slavin BR, Holton T. Comparing Surgical Site Occurrences in 1 versus 2-stage Breast Reconstruction via Federated EMR Network. Plast Reconstr Surg Glob Open. 2021: e3385 [PMID: 33564597 https://doi.org/10.1097/GOX.0000000000003385]
4. Topaloglu U, Palchuk MB. Using a Federated Network of Real-World Data to Optimize Clinical Trials Operations. JCO Clin Cancer Inform. 2018: 1 [PMID: 30652541 https://doi.org/10.1200/CCI.17.00067]
5. Kane LT, Fang T, Galetta MS, Goyal DKC, Nicholson KJ, Kepler CK, Vaccaro AR, Schroeder GD. Propensity Score Matching: A Statistical Method. Clin Spine Surg. 2020: 120 [PMID: 31913173 https://doi.org/10.1097/BSD.0000000000000932]
6. Prasad A, Shin M, Carey RM, Chorath K, Parhar H, Appel S, Moreira A, Rajasekaran K. Propensity score matching in otolaryngologic literature: A systematic review and critical appraisal. PLoS One. 2020: e0244423 [PMID: 33382777 https://doi.org/10.1371/journal.pone.0244423]
7. Badhiwala JH, Karmur BS, Wilson JR. Propensity Score Matching: A Powerful Tool for Analyzing Observational Nonrandomized Data. Clin Spine Surg. 2021: 22 [PMID: 32804684 https://doi.org/10.1097/BSD.0000000000001055]
8. Reiffel JA. Propensity Score Matching: The 'Devil is in the Details' Where More May Be Hidden than You Know. Am J Med. 2020: 178 [PMID: 31618617 https://doi.org/10.1016/j.amjmed.2019.08.055]
9. Mohamed Y, Song X, McMahon TM, Sahil S, Zozus M, Wang Z, Greater Plains C, Waitman LR. Electronic health record data quality variability across a multistate clinical research network. J Clin Transl Sci. 2023: e130 [PMID: 37396818 https://doi.org/10.1017/cts.2023.548]
10. Sandhu N, Whittle S, Varela LO, Eastwood CA, Southern DA, Quan H. Assessing Hospital Data Quality: Application of a Data Quality Tool in 15 Countries. International Journal of Population Data Science. 2024: https://doi.org/10.23889/ijpds.v9i5.2889]
11. London JW, ORourke J, Warnick J, Doole J, De Keyser L, Drebert Z, Wan O, Thompson C, Palchuk M. Deriving breast cancer chemotherapy patterns from real-world data. Journal of Clinical Oncology. 2023: e13586 [PMID: WOS:001053772001478 https://doi.org/10.1200/JCO.2023.41.16_suppl.e13586]
12. Segalas C, Leyrat C, Carpenter JR, Williamson E. Propensity score matching after multiple imputation when a confounder has missing data. Stat Med. 2023: 1082 [PMID: 36695043 https://doi.org/10.1002/sim.9658]
13. Langworthy B, Wu Y, Wang M. An overview of propensity score matching methods for clustered data. Stat Methods Med Res. 2023: 641 [PMID: 36426585 https://doi.org/10.1177/09622802221133556]
14. Wilson JL, Betensky M, Udassi S, Ellison PR, Lilienthal R, Stahl LR, Palchuk MB, Zia A, Town DA, Kimble W, Goldenberg NA, Morizono H. Leveraging a global, federated, real-world data network to optimize investigator-initiated pediatric clinical trials: the TriNetX Pediatric Collaboratory Network. JAMIA Open. 2024: ooae077 [PMID: 39224867 https://doi.org/10.1093/jamiaopen/ooae077]
15. Anand P, Zhang Y, Merola D, Jin Y, Wang SV, Lii J, Liu J, Lin KJ. Comparison of EHR Data-Completeness in Patients with Different Types of Medical Insurance Coverage in the United States. Clin Pharmacol Ther. 2023: 1116 [PMID: 37597260 https://doi.org/10.1002/cpt.3027]
16. Zuo Z, Watson M, Budgen D, Hall R, Kennelly C, Al Moubayed N. Data Anonymization for Pervasive Health Care: Systematic Literature Mapping Study. JMIR Med Inform. 2021: e29871 [PMID: 34652278 https://doi.org/10.2196/29871]
17. Onesimu JA, Karthikeyan J, Eunice J, Pomplun M, Dang H. Privacy Preserving Attribute-Focused Anonymization Scheme for Healthcare Data Publishing. Ieee Access. 2022: 86979 [PMID: WOS:000844128900001 https://doi.org/10.1109/Access.2022.3199433]
18. Talari K, Goyal M. Retrospective studies - utility and caveats. J R Coll Physicians Edinb. 2020: 398 [PMID: 33469615 https://doi.org/10.4997/JRCPE.2020.409]
19. Goldstein ND, Kahal D, Testa K, Burstyn I. Inverse probability weighting for selection bias in a Delaware community health center electronic medical record study of community deprivation and hepatitis C prevalence. Ann Epidemiol. 2021: 1 [PMID: 33933628 https://doi.org/10.1016/j.annepidem.2021.04.011]
20. D'Onofrio BM, Sjolander A, Lahey BB, Lichtenstein P, Oberg AS. Accounting for Confounding in Observational Studies. Annu Rev Clin Psychol. 2020: 25 [PMID: 32384000 https://doi.org/10.1146/annurev-clinpsy-032816-045030]
21. Verbeek JH, Whaley P, Morgan RL, Taylor KW, Rooney AA, Schwingshackl L, Hoving JL, Vittal Katikireddi S, Shea B, Mustafa RA, Murad MH, Schunemann HJ, Group GW. An approach to quantifying the potential importance of residual confounding in systematic reviews of observational studies: A GRADE concept paper. Environ Int. 2021: 106868 [PMID: 34530289 https://doi.org/10.1016/j.envint.2021.106868]
22. Mashoufi M, Ayatollahi H, Khorasani-Zavareh D, Talebi Azad Boni T. Data Quality in Health Care: Main Concepts and Assessment Methodologies. Methods Inf Med. 2023: 5 [PMID: 36716776 https://doi.org/10.1055/s-0043-1761500]
23. Evans L, London JW, Palchuk MB. Assessing real-world medication data completeness. J Biomed Inform. 2021: 103847 [PMID: 34161824 https://doi.org/10.1016/j.jbi.2021.103847]
24. Crutchfield CR, Givens RR, O'Connor M, deMeireles AJ, Lynch TS. Recall Bias in the Retrospective Collection of Common Patient-Reported Outcome Scores in Hip Arthroscopy. Am J Sports Med. 2022: 3190 [PMID: 35993555 https://doi.org/10.1177/03635465221118375]
25. Zini MLL, Banfi G. A Narrative Literature Review of Bias in Collecting Patient Reported Outcomes Measures (PROMs). Int J Environ Res Public Health. 2021: [PMID: 34886170 https://doi.org/10.3390/ijerph182312445]
26. Valentin S, Bramley N, Lucas C. Learning Hidden Causal Structure from Temporal Data. Cognitive Science. 2020: 1906
27. Zhao HH, Shipp AJ, Carter K, Gonzalez-Mulé E, Xu E. Time and change: A meta-analysis of temporal decisions in longitudinal studies. Journal of Organizational Behavior. 2024: 620 [PMID: WOS:001144638500001 https://doi.org/10.1002/job.2771]
28. Kinter S, Delaney JA, Susarla S, McKinney C. Retrospective Cohort Studies in Craniofacial Outcomes Research: An Epidemiologist's Approach to Mitigating Bias. Cleft Palate Craniofac J. 2024: 10556656241233234 [PMID: 38389276 https://doi.org/10.1177/10556656241233234]
29. Degtiar I, Rose S. A Review of Generalizability and Transportability. Annual Review of Statistics and Its Application. 2023: 501 [PMID: WOS:000945740600021 https://doi.org/10.1146/annurev-statistics-042522-103837]
30. Thomas DS, Collin S, Berrocal-Almanza LC, Stirnadel-Farrant H, Zhang Y, Sun P. Extending Inferences From Sample To Target Populations: On The Generalizability Of A Real-World Clinico-Genomic Database Non-Small Cell Lung Cancer Cohort2025: https://doi.org/10.1101/2023.06.15.23291372]
31. Pina E, Ramos J, Jorge H, Váz P, Silva J, Wanzeller C, Abbasi M, Martins P. Data Privacy and Ethical Considerations in Database Management. Journal of Cybersecurity and Privacy. 2024: 494 https://doi.org/10.3390/jcp4030024]
32. Latha Narayanan V, N.Sujatha, Mukul M, Lokesh VS. Ethical considerations in data science: Balancing privacy and utility. International Journal of Science and Research Archive. 2024: 011 https://doi.org/10.30574/ijsra.2024.11.1.1098]
33. Horsky J, Drucker EA, Ramelson HZ. Accuracy and Completeness of Clinical Coding Using ICD-10 for Ambulatory Visits. AMIA Annu Symp Proc. 2017: 912 [PMID: 29854158
34. Chaux R, Treussier I, Audeh B, Pereira S, Hengoat T, Paviot BT, Bousquet C. Automated Control of Codes Accuracy in Case-Mix Databases by Evaluating Coherence with Available Information in the Electronic Health Record. Stud Health Technol Inform. 2019: 551 [PMID: 31437984 https://doi.org/10.3233/SHTI190283]
35. Yu Y, Ruddy KJ, Leventakos K, Liu BL, Huo N, Pachman DR, Zong NS, Xiao GH, Chute C, Pfaff E, Cheville AL, Jiang GQ. Using EHR data and machine learning approach to facilitate the identification of patients with lung cancer from a pan-cancer cohort. Journal of Clinical Oncology. 2023: e13552 [PMID: WOS:001053772001450 https://doi.org/10.1200/JCO.2023.41.16_suppl.e13552]
36. McColm D, Karcz A. Comparing manual and automated coding of physicians quality reporting initiative measures in an ambulatory EHR. J Med Pract Manage. 2010: 6 [PMID: 20839502
37. Luna D, Franco M, Plaza C, Otero C, Wassermann S, Gambarte ML, Giunta D, Gonzalez Bernaldo de Quiros F. Accuracy of an electronic problem list from primary care providers and specialists. Stud Health Technol Inform. 2013: 417 [PMID: 23920588 https://doi.org/10.3233/978-1-61499-289-9-417]
38. Hsu J, Pacheco JA, Stevens WW, Smith ME, Avila PC. Accuracy of phenotyping chronic rhinosinusitis in the electronic health record. Am J Rhinol Allergy. 2014: 140 [PMID: 24717952 https://doi.org/10.2500/ajra.2014.28.4012]
39. Nouraei SA, Hudovsky A, Frampton AE, Mufti U, White NB, Wathen CG, Sandhu GS, Darzi A. A Study of Clinical Coding Accuracy in Surgery: Implications for the Use of Administrative Big Data for Outcomes Management. Ann Surg. 2015: 1096 [PMID: 25470740 https://doi.org/10.1097/SLA.0000000000000851]
40. Olagundoye O, van Boven K, Daramola O, Njoku K, Omosun A. Improving the accuracy of ICD-10 coding of morbidity/mortality data through the introduction of an electronic diagnostic terminology tool at the general hospitals in Lagos, Nigeria. BMJ Open Qual. 2021: [PMID: 33674344 https://doi.org/10.1136/bmjoq-2020-000938]
41. Yordanov TR, Abu-Hanna A, Ravelli ACJ, Vagliano I. Autoencoder-Based Prediction of ICU Clinical Codes. Artificial Intelligence in Medicine, Aime 2023. 2023: 57 [PMID: WOS:001295128100007 https://doi.org/10.1007/978-3-031-34344-5_8]
42. Pellathy T, Saul M, Clermont G, Dubrawski AW, Pinsky MR, Hravnak M. Accuracy of identifying hospital acquired venous thromboembolism by administrative coding: implications for big data and machine learning research. J Clin Monit Comput. 2022: 397 [PMID: 33558981 https://doi.org/10.1007/s10877-021-00664-6]
43. Schrodi SJ. The Impact of Diagnostic Code Misclassification on Optimizing the Experimental Design of Genetic Association Studies. J Healthc Eng. 2017: 7653071 [PMID: 29181145 https://doi.org/10.1155/2017/7653071]
44. Farzandipour M, Sheikhtaheri A. Accuracy of diagnostic coding based on ICD-10. KAUMS Journal. 2009: 67
45. Usher M, Sahni N, Herrigel D, Simon G, Melton GB, Joseph A, Olson A. Diagnostic Discordance, Health Information Exchange, and Inter-Hospital Transfer Outcomes: a Population Study. J Gen Intern Med. 2018: 1447 [PMID: 29845466 https://doi.org/10.1007/s11606-018-4491-x]
46. Zafirah SA, Nur AM, Puteh SEW, Aljunid SM. Potential loss of revenue due to errors in clinical coding during the implementation of the Malaysia diagnosis related group (MY-DRG((R))) Casemix system in a teaching hospital in Malaysia. BMC Health Serv Res. 2018: 38 [PMID: 29370785 https://doi.org/10.1186/s12913-018-2843-1]
47. Lorence DP, Ibrahim IA. Benchmarking variation in coding accuracy across the United States. J Health Care Finance. 2003: 29 [PMID: 12908652
48. Re V. Validation of health outcomes of interest in healthcare databases2021: 207 https://doi.org/10.1016/B978-0-12-817663-4.00022-2]
49. Palestine AG, Merrill PT, Saleem SM, Jabs DA, Thorne JE. Assessing the Precision of ICD-10 Codes for Uveitis in 2 Electronic Health Record Systems. JAMA Ophthalmol. 2018: 1186 [PMID: 30054618 https://doi.org/10.1001/jamaophthalmol.2018.3001]
50. Stewart CC, Lu CY, Yoon TK, Coleman KJ, Crawford PM, Lakoma MD, Simon GE. Impact of ICD-10-CM Transition on Mental Health Diagnoses Recording. EGEMS (Wash DC). 2019: 14 [PMID: 31065557 https://doi.org/10.5334/egems.281]
51. Quan H, Sundararajan V, Halfon P, Fong A, Burnand B, Luthi JC, Saunders LD, Beck CA, Feasby TE, Ghali WA. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care. 2005: 1130 [PMID: 16224307 https://doi.org/10.1097/01.mlr.0000182534.19832.83]
52. Caskey R, Zaman J, Nam H, Chae SR, Williams L, Mathew G, Burton M, Li JJ, Lussier YA, Boyd AD. The transition to ICD-10-CM: challenges for pediatric practice. Pediatrics. 2014: 31 [PMID: 24918217 https://doi.org/10.1542/peds.2013-4147]
53. Gonzalez L, Perez-Rey D, Alonso E, Hernandez G, Serrano P, Pedrera M, Gomez A, De Schepper K, Crepain T, Claerhout B. Building an I2B2-Based Population Repository for Clinical Research. Stud Health Technol Inform. 2020: 78 [PMID: 32570350 https://doi.org/10.3233/SHTI200126]
54. Haudenschild C, Vaickus L, Levy J. Configuring a federated network of real-world patient health data for multimodal deep learning prediction of health outcomes. bioRxiv. 2021: https://doi.org/10.1145/3477314.3507007]
55. Torner A, Dickman P, Duberg AS, Kristinsson S, Landgren O, Bjorkholm M, Svensson A. A method to visualize and adjust for selection bias in prevalent cohort studies. Am J Epidemiol. 2011: 969 [PMID: 21920949 https://doi.org/10.1093/aje/kwr211]
56. Gkatzionis A, Burgess S. Contextualizing selection bias in Mendelian randomization: how bad is it likely to be? Int J Epidemiol. 2019: 691 [PMID: 30325422 https://doi.org/10.1093/ije/dyy202]
57. Grimes DA, Schulz KF. Bias and causal associations in observational research. Lancet. 2002: 248 [PMID: 11812579 https://doi.org/10.1016/S0140-6736(02)07451-2]
58. Haneuse S. Distinguishing Selection Bias and Confounding Bias in Comparative Effectiveness Research. Med Care. 2016: e23 [PMID: 24309675 https://doi.org/10.1097/MLR.0000000000000011]
59. Valeri L, Coull BA. Estimating causal contrasts involving intermediate variables in the presence of selection bias. Stat Med. 2016: 4779 [PMID: 27411847 https://doi.org/10.1002/sim.7025]
60. Bateson TF, Schwartz J. Selection bias and confounding in case-crossover analyses of environmental time-series data. Epidemiology. 2001: 654 [PMID: 11679793 https://doi.org/10.1097/00001648-200111000-00013]
61. Infante-Rivard C, Cusson A. Reflection on modern methods: selection bias-a review of recent developments. Int J Epidemiol. 2018: 1714 [PMID: 29982600 https://doi.org/10.1093/ije/dyy138]
62. Howe CJ, Cole SR, Lau B, Napravnik S, Eron JJ, Jr. Selection Bias Due to Loss to Follow Up in Cohort Studies. Epidemiology. 2016: 91 [PMID: 26484424 https://doi.org/10.1097/EDE.0000000000000409]
63. Kim TH, Samson LF, Lu N. Racial/ethnic disparities in the utilization of high-technology hospitals. J Natl Med Assoc. 2010: 803 [PMID: 20922924 https://doi.org/10.1016/s0027-9684(15)30677-5]
64. Groeneveld PW, Laufer SB, Garber AM. Technology diffusion, hospital variation, and racial disparities among elderly Medicare beneficiaries: 1989-2000. Med Care. 2005: 320 [PMID: 15778635 https://doi.org/10.1097/01.mlr.0000156849.15166.ec]
65. Sequist TD. Health information technology and disparities in quality of care. J Gen Intern Med. 2011: 1084 [PMID: 21809173 https://doi.org/10.1007/s11606-011-1812-8]
66. David Y, Jahnke EG. Planning Medical Technology Management in a Hospital. Global Clinical Engineering Journal. 2018: 23 https://doi.org/10.31354/globalce.v0i1.23]
67. Newton RC, Mytton OT, Aggarwal R, Runciman WB, Free M, Fahlgren B, Akiyama M, Farlow B, Yaron S, Locke G, Whittaker S. Making existing technology safer in healthcare. Qual Saf Health Care. 2010: i15 [PMID: 20693212 https://doi.org/10.1136/qshc.2009.038539]
68. Walker DM, Hefner JL, Fareed N, Huerta TR, McAlearney AS. Exploring the Digital Divide: Age and Race Disparities in Use of an Inpatient Portal. Telemed J E Health. 2020: 603 [PMID: 31313977 https://doi.org/10.1089/tmj.2019.0065]
69. Chin MH, Clarke AR, Nocon RS, Casey AA, Goddu AP, Keesecker NM, Cook SC. A roadmap and best practices for organizations to reduce racial and ethnic disparities in health care. J Gen Intern Med. 2012: 992 [PMID: 22798211 https://doi.org/10.1007/s11606-012-2082-9]
70. Kilbourne AM, Switzer G, Hyman K, Crowley-Matoka M, Fine MJ. Advancing health disparities research within the health care system: a conceptual framework. Am J Public Health. 2006: 2113 [PMID: 17077411 https://doi.org/10.2105/AJPH.2005.077628]
71. Rust G, Cooper LA. How can practice-based research contribute to the elimination of health disparities? J Am Board Fam Med. 2007: 105 [PMID: 17341746 https://doi.org/10.3122/jabfm.2007.02.060131]
72. Chinman M, Woodward EN, Curran GM, Hausmann LRM. Harnessing Implementation Science to Increase the Impact of Health Equity Research. Med Care. 2017: S16 [PMID: 28806362 https://doi.org/10.1097/MLR.0000000000000769]
73. Koh HK, Oppenheimer SC, Massin-Short SB, Emmons KM, Geller AC, Viswanath K. Translating research evidence into practice to reduce health disparities: a social determinants approach. Am J Public Health. 2010: S72 [PMID: 20147686 https://doi.org/10.2105/AJPH.2009.167353]
74. Lane M, Bell R, Latham-Sadler B, Bradley C, Foxworth J, Smith N, Millar A, Hairston K, Roper B, Howlett A. Translational Research Training at Various Levels of Professional Experience to Address Health Disparities. Journal of best practices in health professions diversity : research, education and policy. 2016: 1178
75. Dankwa-Mullan I, Rhee KB, Stoff DM, Pohlhaus JR, Sy FS, Stinson N, Jr., Ruffin J. Moving toward paradigm-shifting research in health disparities through translational, transformational, and transdisciplinary approaches. Am J Public Health. 2010: S19 [PMID: 20147662 https://doi.org/10.2105/AJPH.2009.189167]
76. Baker EL, White LE, Lichtveld MY. Reducing health disparities through community-based research. Public Health Reports. 2001: 517 [PMID: WOS:000177607300004 https://doi.org/10.1016/s0033-3549(04)50083-3]
77. Stuart EA, Bradshaw CP, Leaf PJ. Assessing the generalizability of randomized trial results to target populations. Prev Sci. 2015: 475 [PMID: 25307417 https://doi.org/10.1007/s11121-014-0513-z]
78. Murad MH, Katabi A, Benkhadra R, Montori VM. External validity, generalisability, applicability and directness: a brief primer. BMJ Evid Based Med. 2018: 17 [PMID: 29367319 https://doi.org/10.1136/ebmed-2017-110800]
79. Pearl J, Bareinboim E. External Validity: From Do-Calculus to Transportability Across Populations. Statistical Science. 2014: 579 [PMID: WOS:000348786900006 https://doi.org/10.1214/14-Sts486]
80. Leviton LC. Generalizing about Public Health Interventions: A Mixed-Methods Approach to External Validity. Annu Rev Public Health. 2017: 371 [PMID: 28125391 https://doi.org/10.1146/annurev-publhealth-031816-044509]
81. Ling AY, Montez-Rath ME, Carita P, Chandross KJ, Lucats L, Meng Z, Sebastien B, Kapphahn K, Desai M. An Overview of Current Methods for Real-world Applications to Generalize or Transport Clinical Trial Findings to Target Populations of Interest. Epidemiology. 2023: 627 [PMID: 37255252 https://doi.org/10.1097/EDE.0000000000001633]
82. Kennedy-Martin T, Curtis S, Faries D, Robinson S, Johnston J. A literature review on the representativeness of randomized controlled trial samples and implications for the external validity of trial results. Trials. 2015: 495 [PMID: 26530985 https://doi.org/10.1186/s13063-015-1023-4]
83. Evans D. Hierarchy of evidence: a framework for ranking evidence evaluating healthcare interventions. J Clin Nurs. 2003: 77 [PMID: 12519253 https://doi.org/10.1046/j.1365-2702.2003.00662.x]
84. Hernandez S, Fairchild K, Pemberton M, Dahmer J, Zhang W, Palchuk MB, Topaloglu U. Applying FHIR Genomics for Research - From Sequencing to Database. AMIA Jt Summits Transl Sci Proc. 2022: 236 [PMID: 35854733
85. Evans L, London JW, Palchuk MB. The Detection of Date Shifting in Real-World Data. Appl Clin Inform. 2023: 763 [PMID: 37459888 https://doi.org/10.1055/a-2130-2197]
86. Antoine A, Perol D, Robain M, Delaloge S, Lasset C, Drouet Y. Target trial emulation to assess real-world efficacy in the Epidemiological Strategy and Medical Economics metastatic breast cancer cohort. J Natl Cancer Inst. 2023: 971 [PMID: 37220893 https://doi.org/10.1093/jnci/djad092]
87. Furler J, Magin P, Pirotta M, van Driel M. Participant demographics reported in "Table 1" of randomised controlled trials: a case of "inverse evidence"? Int J Equity Health. 2012: 14 [PMID: 22429574 https://doi.org/10.1186/1475-9276-11-14]
88. Munoz Monjas A, Rubio Ruiz D, Perez-Rey D, Palchuk M. Automatic Outlier Detection in Laboratory Result Distributions Within a Real World Data Network. Stud Health Technol Inform. 2023: 88 [PMID: 37203615 https://doi.org/10.3233/SHTI230070]
89. Enewold L, Parsons H, Zhao L, Bott D, Rivera DR, Barrett MJ, Virnig BA, Warren JL. Updated Overview of the SEER-Medicare Data: Enhanced Content and Applications. J Natl Cancer Inst Monogr. 2020: 3 [PMID: 32412076 https://doi.org/10.1093/jncimonographs/lgz029]
90. Forrest CB, McTigue KM, Hernandez AF, Cohen LW, Cruz H, Haynes K, Kaushal R, Kho AN, Marsolo KA, Nair VP, Platt R, Puro JE, Rothman RL, Shenkman EA, Waitman LR, Williams NA, Carton TW. PCORnet(R) 2020: current state, accomplishments, and future directions. J Clin Epidemiol. 2021: 60 [PMID: 33002635 https://doi.org/10.1016/j.jclinepi.2020.09.036]
91. Qualls LG, Phillips TA, Hammill BG, Topping J, Louzao DM, Brown JS, Curtis LH, Marsolo K. Evaluating Foundational Data Quality in the National Patient-Centered Clinical Research Network (PCORnet(R)). EGEMS (Wash DC). 2018: 3 [PMID: 29881761 https://doi.org/10.5334/egems.199]
92. Ganslandt T, Mate S, Helbing K, Sax U, Prokosch HU. Unlocking Data for Clinical Research - The German i2b2 Experience. Appl Clin Inform. 2011: 116 [PMID: 23616864 https://doi.org/10.4338/ACI-2010-09-CR-0051]
93. Feroz AS, Hussaini AS, Seto E. Feasibility and Ethical Considerations for Conducting Online versus In-person Interviews for a Qualitative Study. Preventive Medicine: Research & Reviews. 2024: 321 https://doi.org/10.4103/pmrr.Pmrr_197_24]
94. Adnyana IMDM, Utomo B, Eljatin DS, Setyawan MF. Developing and Establishing Attribute-based Surveillance System: A Review. Preventive Medicine: Research & Reviews. 2024: 76 https://doi.org/10.4103/pmrr.Pmrr_54_23]
95. Karol S, Thakare MM. Strengthening Immunisation Services in India through Digital Transformation from Co-WIN to U-WIN: A Review. Preventive Medicine: Research & Reviews. 2024: 25 https://doi.org/10.4103/pmrr.Pmrr_18_23]

This work is licensed under a Creative Commons Attribution 4.0 International License.
Copyright (c) 2025 Mahmoud Nassar, Hazem Abosheaishaa, Khaled Elfert, Azizullah Beran, Abdellatif Ismail, Mouhand Mohamed, Anoop Misra, Muhammed Amir Essibayi, David J. Altschul, Ahmed Y. Azzam